Software tools
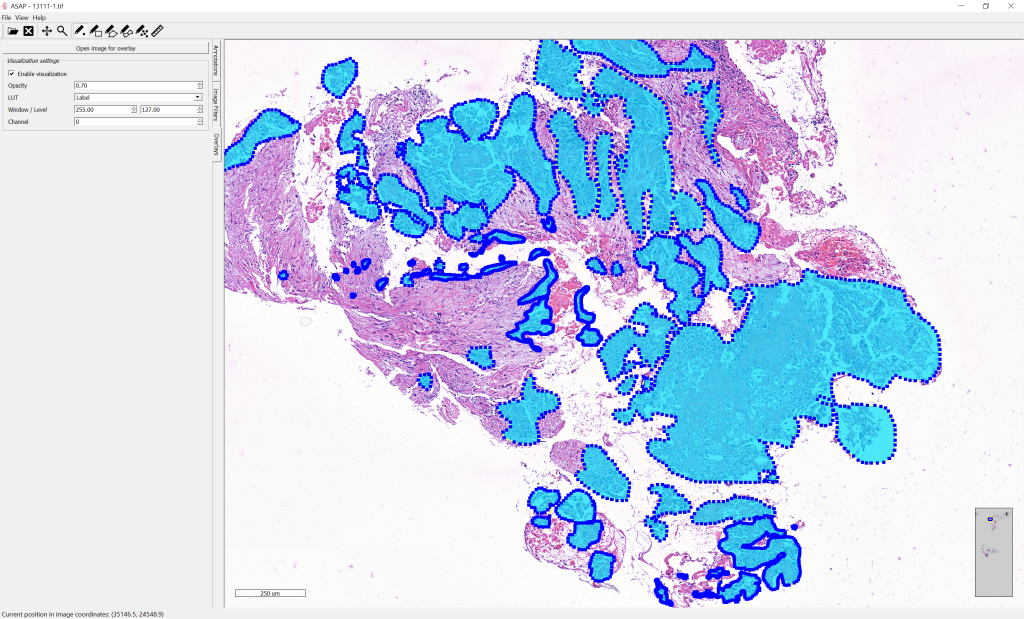
Visualizing Whole-Slide Images and Annotations
Reading the multi-resolution images using standard image tools or libraries is a challenge because these tools are typically designed for images that can comfortably be uncompressed into RAM or a swap file. OpenSlide is a C library that provides a simple interface to read WSIs of different formats.
Automated Slide Analysis Platform (ASAP) is an open source platform developed by the Computational Pathology Group for visualising, annotating and automatically analysing whole-slide histopathology images. ASAP is built on top of several well-developed open source packages like OpenSlide, Qt and OpenCV. We strongly recommend the participants to use this platform for visualising the slides and viewing the annotations.
Accessing the data
The whole-slide images provided in this challenge are standard TIFF files. Standard libraries like OpenSlide can be used to open and read these files. We used ASAP to prepare the data. Its multiresolutionimageinterface C++ library and python package provides an easy to use interface for accessing pixel data in TIFF files efficiently. Only 3 simple steps are necessary to be able to use it in python:
- Download and install ASAP.
- Configure your PYTHONPATH environment variable to contain the <ASAP install directory>/bin directory path.
- Import multiresolutionimageinterface to your python module.
A few things to know about the library and the TIFF image format:
- The TIFF contains multiple down-sampled versions of the original image. The highest resolution in on level 0.
- Pixel values can be read in patches from any available level.
- Pixel indexing is done in (column, row) manner.
- Regardless of which level a patch is read from the indexing is done on level 0.
The following python code snippet loads a TIFF file and reads a 300 pixel wide, 200 pixel high image patch starting at the (568, 732) XY coordinate on level 2:
import multiresolutionimageinterface as mir
reader = mir.MultiResolutionImageReader()
mr_image = reader.open('ACDC_challenge/Images/13111-1.tif')
level = 2
ds = mr_image.getLevelDownsample(level)
image_patch = mr_image.getUCharPatch(int(568 * ds), int(732 * ds), 300, 200, level)
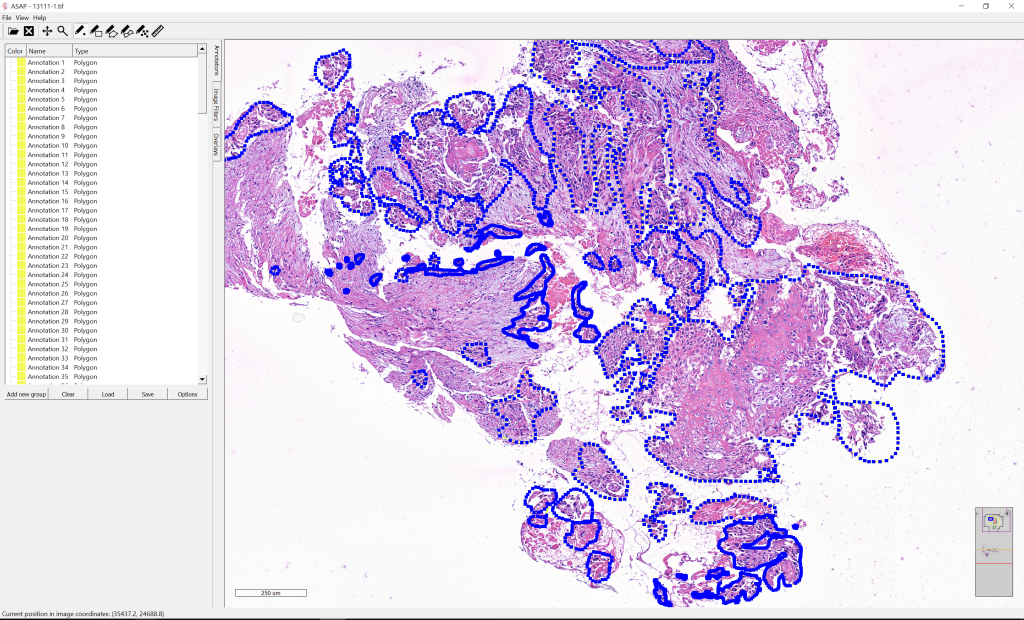
Annotations
The annotations were made in ASAP. The annotation ROIs are polygons that are stored as an ordered list of vertex (X, Y) pixel coordinates on level 0 of the multi-resolution images.
The following python code snippet converts an annotation into a mask file:
import multiresolutionimageinterface as mir
reader = mir.MultiResolutionImageReader()
mr_image = reader.open('ACDC_challenge/Images/13111-1.tif')
annotation_list = mir.AnnotationList()
xml_repository = mir.XmlRepository(annotation_list)
xml_repository.setSource('ACDC_challenge/Annotations/13111-1.xml')
xml_repository.load()
annotation_mask = mir.AnnotationToMask()
output_path = 'ACDC_challenge/Images/13111-1_tumor_annotations.tif'
annotation_mask.convert(annotation_list, output_path, mr_image.getDimensions(), mr_image.getSpacing())After conversion, the mask of manual annotations can be visualized in overlay on top of the whole-slide image: